![]()
Guangyuan(Frank) Li
Biomedical Informatics first-year PhD student
University of Cincinnati
Cincinnati Children Hospital Medical Center(CCHMC)
Email: li2g2@mail.uc.edu
Education
09/2018 – 05/2019
Exchange student in Biological Design Institute, Arizona State University, U.S.A
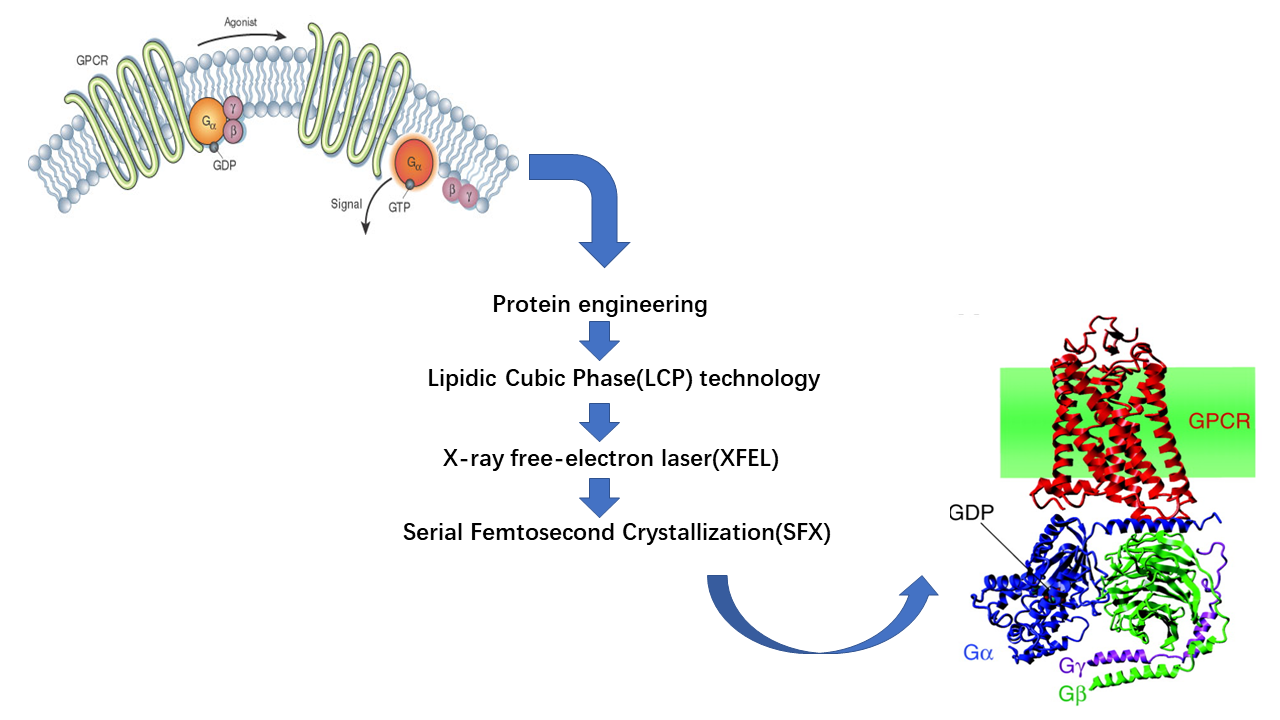
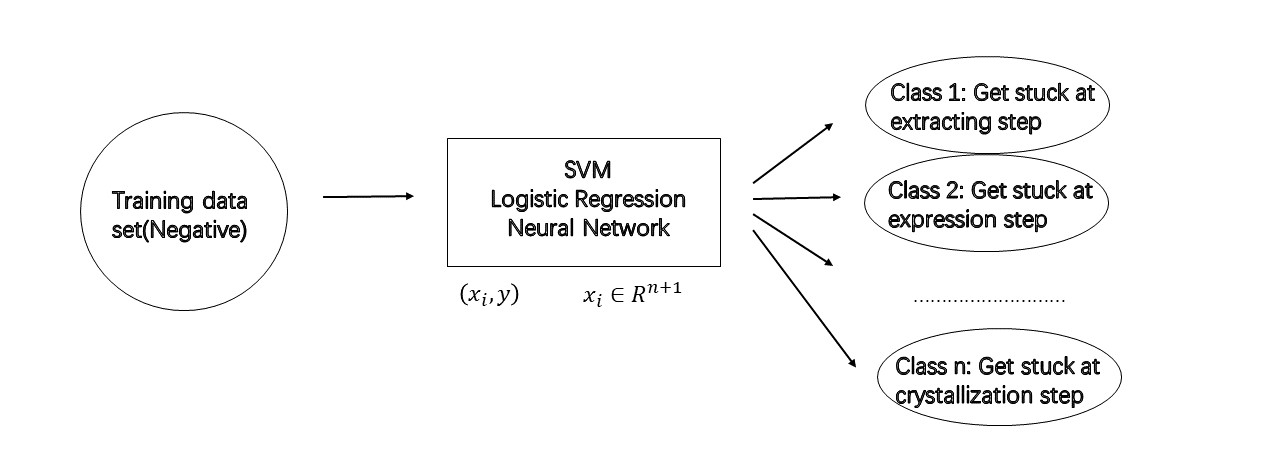
Project: Analyzing GPCR structure by using lipidic phase technology (Supervisor: Dr. Wei Liu)
Click the link to know more about my GPCR project: GPCR project
09/2015 – 06/2019
Bachelor of biology in college of life sciences, Wuhan university, China
General GPA: 3.81/4.00
Courses: Biology category, Computer Science category, Mathematical category, etc.
Click each link will redirect to my course lists
Click the link to know more about my home institution: Wuhan University
Research experience
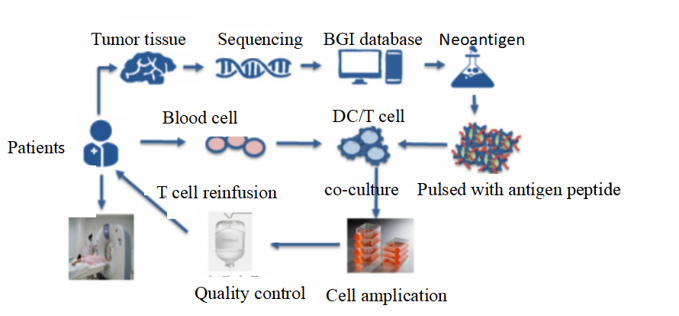
Shenzhen Huada Gene Research Institute, BGI-shenzhen, March 2018-July 2018
Intern, An novel cell therapy for melonoma and colon cancer
- Participated in drug pre-clinical testing and drug declaration process
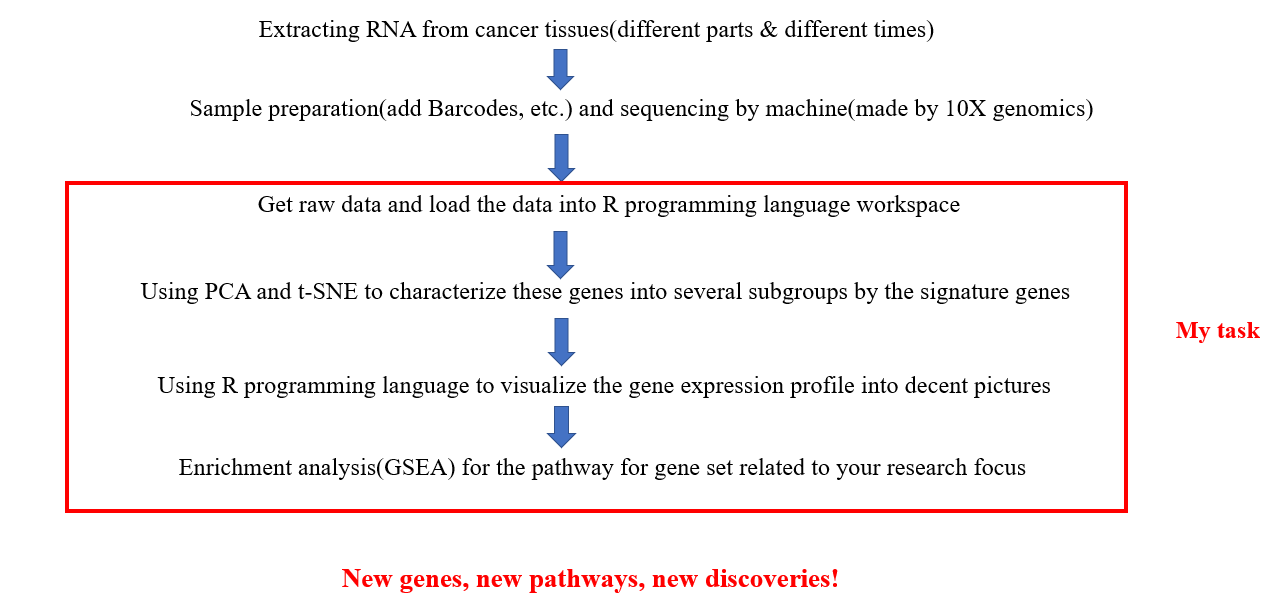
- Used R programming language to analyze single cell RNA-seq data
- Took systematic bioinformatic training(Linux, Python, Perl, R programming language )
- Familiar with the cutting-edge progress of cancer immunotherapy
Click on the link to know more: BGI internship
College of life science, Wuhan University, November 2016-November 2017
Core member, iGEM(international genetically engineered machine) competition
- Won silver medal in the final presentation(Boston,USA)
- Designed an engineered bacteria for disposing of sewage
- In charge of mathematical modelling-building and performance testing
- Took part in experimental part including molecular cloning, cell culture, etc
Click on the link to know more:iGEM journal
Institute of hydrobiology, CAS, May 2017-May 2018
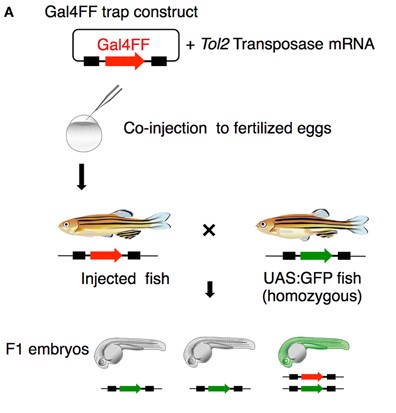
Research assistant, Early development of zebrafish Primodial Germ Cell(PGC)
- Studied the function of several critical genes involved in zebrafish PGC cell formation and sex determination
- Conducted zebrafish genetic experiment, including microinjection, in-situ hybridization, confocal microscopy, immunofluorescence
Click on the link to know more: zebrafish research
Highlights
- Having systematic bioinformatic training
- Familiar in Perl, Python, R computer programming languages and MATLAB, can solve personalized programming demands
- Structural biology research experience
- Molecular cloning technology, cell culture operation, genetic operation in zebrafish
- Good at communicating with others
- Having high team spirits
Honors & Scholarships
- Excellent student in Wuhan University(Three consecutive years) 2016,2017,2018
- Excellent scholarship in Wuhan University(Two consecutive years) 2016,2017
- Best debater in Wuhan University debating tournament
- Vice director in department student union