A novel set of cancer therapy
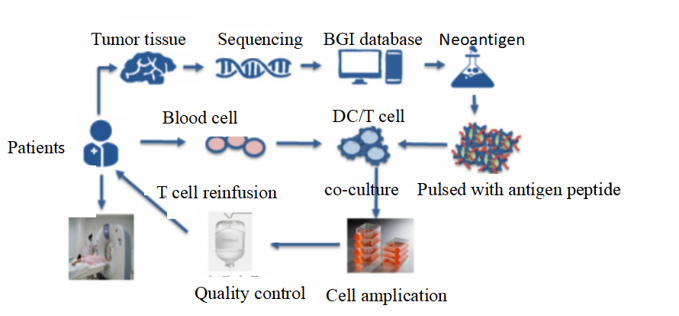
I am now working as an intern in BGI-Shenzhen. BGI is one of the world’s genome sequencing center, my job is to participate in developing a novel anti-tumor method called TSA-CTL. Based upon the powerful and accurate sequencing platform, we can firstly identify tumor specific antigen(TSA) by using whole exome sequencing(WES) and RNA-seq, and use these antigens to activate patients’ self-derived cytotoxic lymphocyte(CTL) in vitro, finally reinfuse the activated T cell into patients’ body. 2 out of 5 patients of malignant melanoma have appeared partial tumor regression during our clinical trial. More importantly, different from CAR-T therapy, our method is free from off-target effect or cytokine syndrome, all of our clinical trials are reported to be safe.
The biggest harvest for me is the systematic bioinformatic training, ranging from sequencing technology, processing single-cell RNA data, to learning python, R,and perl to solve series of programming probelm. And I am currently learning how to make good use of the TCGA(The Cancer Genome Atlas) database to do data mining and bioinformatic analysis like survival analysis and gene expression profile.
Single cell RNA-seq analysis
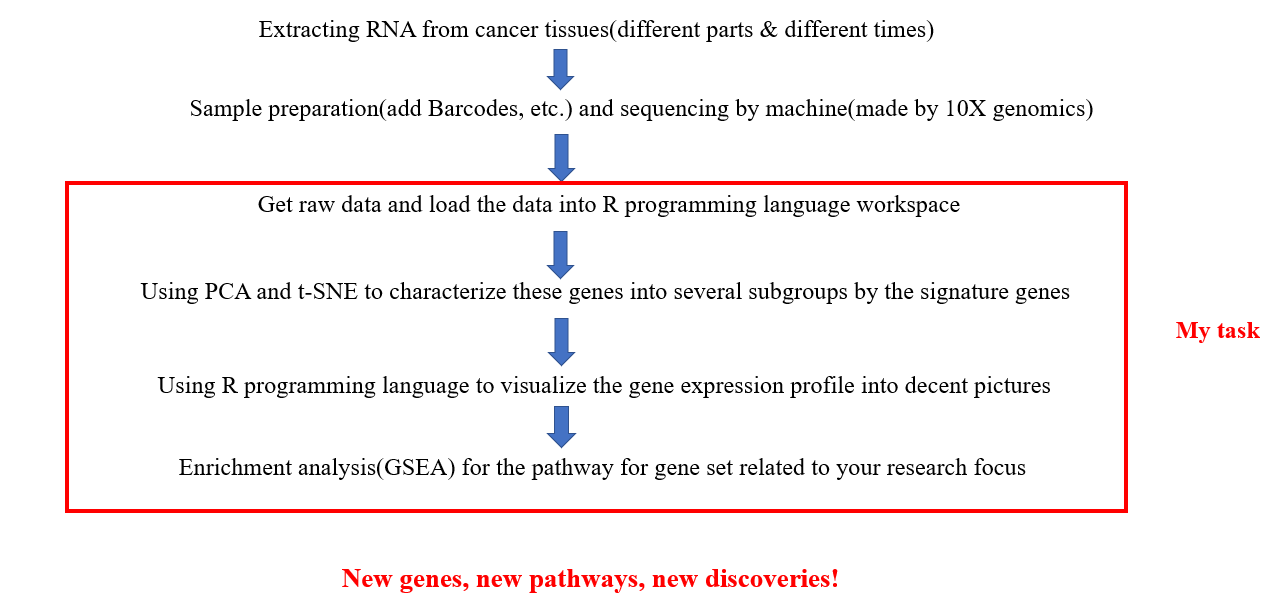
By using single cell RNA-seq, we aim to detect those unknown critical genes in cancer development. Plus, we can compare the gene expression profile in different area and different time, we can track the effects of a certain kind of drug and the whole developmental timeline. Combined with enrichment analysis, we can determine which set of gene would be most correlated with our observed phenotype.
Training program ( Bulk RNA-seq analysis)
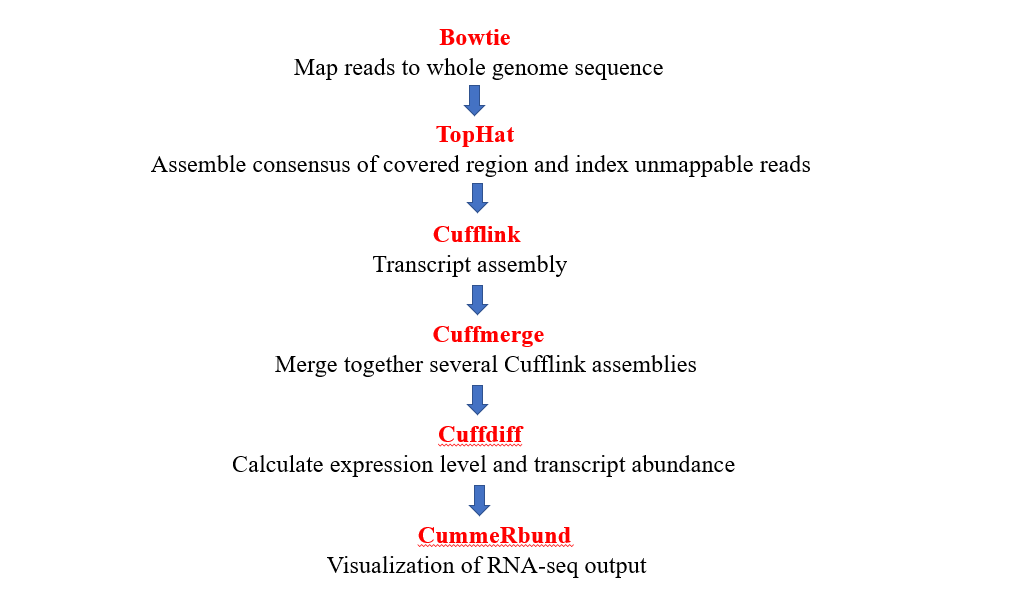
Analyzing bulk RNA-seq data is the most basic and easy-to-learn pipeline in bioinformatics analysis. We chose Bowtie to map and align our RNA reads, then TopHat came up to fix the “junction site” problem, which is caused by alternative splicing. Cufflink is a basic package that can help us calculate gene expression, along with analyzing significant diffrences between two or more samples. Finally,we deployed R package to visulize the results.